1、分层聚类
#载入R自带的测试数据
#data(iris)
#attach(iris)
inData = iris[,1:4]
#计算距离矩阵,并绘图
inData.dist = dist(inData)
#inData.dist = dist(inData,method='euclidean')
heatmap(as.matrix(inData.dist), labRow = F, labCol = F)
#进行分层聚类
#并绘图
#inData.hc <- hclust(inData.dist)
#inData.hc <- hclust(inData.dist,method='ward')
plot(inData.hc, labels = FALSE, hang = -1)
#标识聚类结果,结果设为3类
rect.hclust(inData.hc, k = 3)
#将Tree进行分组
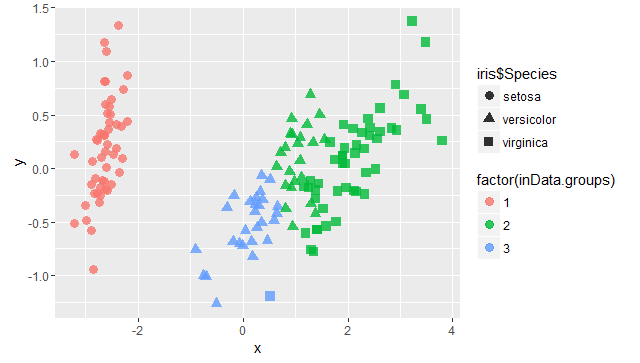
inData.groups <- cutree(inData.hc, 3)
#输出结果表格
table(inData.groups, Species)
#进行降维处理
#绘图对比结果
#形状是正确的数据
#颜色为聚类后的数据
mds=cmdscale(inData.dist,k=2,eig=T)
x = mds$points[,1]
y = mds$points[,2]
library(ggplot2)
p=ggplot(data.frame(x,y),aes(x,y))
p+geom_point(size=3,alpha=0.8,aes(colour=factor(inData.groups),shape=iris$Species))
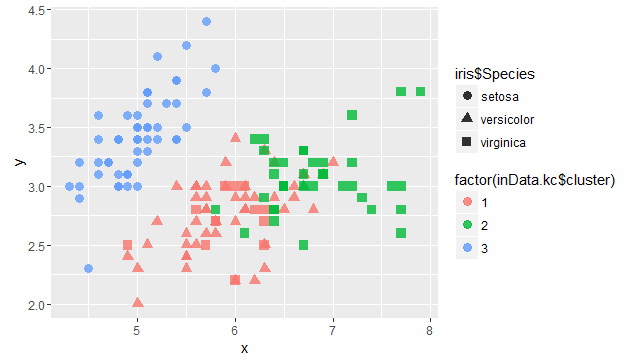
2、K值聚类
#载入R自带的测试数据
#data(iris)
#attach(iris)
inData = iris[,1:4]
#计算距离矩阵,并绘图
inData.dist = dist(inData)
#inData.dist = dist(inData,method='euclidean')
heatmap(as.matrix(inData.dist), labRow = F, labCol = F)
#进行K值聚类
inData.kc <- kmeans(inData.dist,centers=3)
#绘图对比结果
#形状是正确的数据
#颜色为聚类后的数据
library(ggplot2)
x=inData[c("Sepal.Length")]
y=inData[c("Sepal.Width")]
p=ggplot(data.frame(x,y),aes(x,y))
p+geom_point(size=3,alpha=0.8,aes(colour=factor(inData.kc$cluster),shape=iris$Species))